

|
EIDORS: Electrical Impedance Tomography and Diffuse Optical Tomography Reconstruction Software |
|
EIDORS
(mirror) Main Documentation Tutorials − Image Reconst − Data Structures − Applications − FEM Modelling − GREIT − Old tutorials − Workshop Download Contrib Data GREIT Browse Docs Browse SVN News Mailing list (archive) FAQ Developer
|
GREIT Reconstruction for multiple electrode layersThe GREIT formulation can be directly used to reconstruct a single plane in the centre of an EIT configuration in which two (or more) planes of electrodes are used.Simulation model
% $Id: mk_GREIT_mat_2layer01.m 2554 2011-04-28 19:14:20Z aadler $
n_elecs = 8;
layers = [0.8,1.2];
stim = mk_stim_patterns(n_elecs*length(layers),1,[0,1],[0,1], ...
{'no_meas_current'}, 1);
extra={'lungs','solid lungs = sphere(0.9,0.1,1.65;0.3) or sphere(-0.9,0.1,1;0.3);'};
[fmdl,midx] = ng_mk_ellip_models([2, 2,1.4,0.2] ,[n_elecs,layers],[0.1], extra);
fmdl.stimulation = stim;
img = mk_image(fmdl,1); % Homogeneous background
vh = fwd_solve(img);
img.elem_data(midx{2}) = 0.5; % Lung regions
vi = fwd_solve(img);
show_fem(img,[0,1]); view(0,70);
print_convert mk_GREIT_mat_2layer01a.png '-density 60'
view(0,10);
print_convert mk_GREIT_mat_2layer01b.png '-density 60'
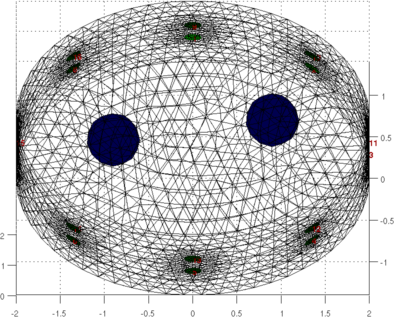

Figure: Simulation of data on an elliptical model with lung shaped contrasting regions Calculate GREIT reconstruction matrix% $Id: mk_GREIT_mat_2layer02.m 4341 2013-09-24 16:08:49Z aadler $ opt.imgsz = [32 32]; opt.distr = 3; % non-random, uniform opt.Nsim = 1000; opt.target_size = 0.05; % Target size (frac of medium) opt.noise_figure = 1.0; % Recommended NF=0.5; % opt.target_layer = 1.0; % Default is in the centre fmdl = ng_mk_ellip_models([2, 2,1.4,0.2] ,[n_elecs,layers],[0.1]); fmdl.stimulation = stim; fmdl = mdl_normalize(fmdl, 0); img = mk_image(fmdl,1); imdl = mk_GREIT_model(img, 0.25, [], opt); show_fem(fmdl); view(0,70); print_convert mk_GREIT_mat_2layer02a.png '-density 60' 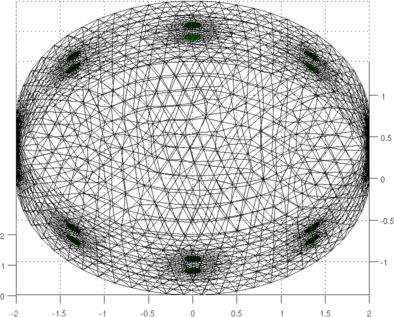
Figure: Simulation of data on an elliptical model with lung shaped contrasting regions Reconstruct Images% $Id: mk_GREIT_mat_2layer03.m 2553 2011-04-28 19:03:58Z aadler $ rimg = inv_solve(imdl,vh,vi); %Reconstruct rimg.calc_colours.ref_level=0; show_fem(rimg); axis square; print_convert mk_GREIT_mat_2layer03a.png '-density 60' 
Figure: Reconstructed images |
Last Modified: $Date: 2017-02-28 13:12:08 -0500 (Tue, 28 Feb 2017) $ by $Author: aadler $